And in the past 50 years, science has discovered that at the very foundation of life there are sophisticated molecular machines, which do the work in the cell. I mean, literally, there are real machines inside everybody’s cells and this is what they are called by all biologists who work in the field, molecular machines. They’re little trucks and busses that run around the cell that takes supplies from one end of the cell to the other. They’re little traffic signals to regulate the flow. They’re sign posts to tell them when they get to the right destination. They’re little outboard motors that allow some cells to swim. If you look at the parts of these, they’re remarkably like the machineries that we use in our everyday world.
The argument is that we know from experience that machinery in our everyday world that we use in our everyday world required design, required an intelligent agent that put it together, who understood how it was going to be used and who assembled the parts. By an inductive argument, when we find such sophisticated machinery in other places too, we can conclude that it also requires design. So now that we found it in life and in the very foundation of life, I and other ID advocates argue that there is no reason to not reach the same conclusion and that in fact, these things were indeed designed.
Actually the correct conclusion would be that since all the machines we are familiar with were designed by humans, cellular machines were designed by humans as well - but note the shifting language of the premise.
Also here Behe says:
In many biological structures proteins are simply components of larger molecular machines. Like the picture tube, wires, metal bolts and screws that comprise a television set, many proteins are part of structures that only function when virtually all of the components have been assembled.
And here is Calvert:
We intuitively infer design when we observe the awesome complexity of the living information processing systems that comprise life. When we look into the black box of the cell we see a biological language, input devices, application programs, information processors, output devices, and systems designed to collect and process energy, make decisions and direct the construction, maintenance and operation of cellular machines and systems. DNA is a blue print that has a semantic or meaningful characteristic found in any writing produced by a mind. That semantic characteristic has not been explained by natural law and chance. Non-natural machines and information processing systems are the kinds of effects that are produced only by human minds. Hence, analogy leads to a reasonable inference that intelligence may also be the cause of similar biological machines and information processing systems.
So, how acurate is the analogy? Let's look at protein synthesis. Before proceeding, however, let me mention that the following synopsis is drawn from Genes VI by Benjamin Lewin and Molecular Biology of the Cell by Alberts et al. Any mistakes (and I hope there are few) are mine.
Ribosomes


Anyone who has had biology knows the process (the picture above is a graphic overview of the process). The DNA helix unwinds, the mRNA makes a complementary template, ribosomes bind to the mRNA strand and tRNA brings the amino acids that will form the protein. This is the way it is usually presented (with a little more detail) and it all sounds very straight forward and machine like. In reality, there is a lot more to the process than that. In bacteria free RNA polymerase collide randomly with DNA , with most sticking very weakly to the DNA. When the RNA polymerase hits the promotor region, however, it binds tightly and transcription begins (which in and of itself is a good argument against the whole notion of irreducible complexity - which is related to the machine analogy).
In eucaryotic cells (cells with a nucleus) the transcription part of the process occurs in the nucleus. The DNA strands are opened by an RNA polymerase which exposes a short section of nucleotides (I'm still simplifying greatly) . The RNA polymerase then binds incoming ribonucleoside triphosphate monomers to form a chain. Once a RNA chain is completed it moves out in the cytoplasm for the translation process. It is here where I really want to pick up the story in more detail.
In order for the translation process to occur the mRNA molecule has to be unwound and there are several factors ( called, in eucaryotic cells, eIF-4, eIF-4A and eIF-4B) that actually start unwinding the mRNA molecule at the 5' end.
mRNA


Ribosomes then attach to the 5' end and migrate towards the initiator codon - a sequence of the three bases AUG. The distance between the 5' end and the initiator codon varies between 50 and 1000 bases (imagine if the timing chain on your car displayed similar variation in where it was set - while the car was running, would it still run?). As it moves down the mRNA the ribosome occasionally encounters obstacles - called secondary structure hairpins - that prevent it from moving further. At that point the ribosome disassociates into it's two smaller subunits and the process starts over again (imagine if you car had a blocked gas line - could it over come that and continue running?). The optimum start sequence is GCCA/GCAUGG. The A or G three sequences before the start codon and the G immediately after are particularly important, they increase the efficiency of translation by a factor of 10 (which would be like having a car that got 10 mpg when you put regular in it but 100 mpg when you put unleaded).
tRNA Cloverleaf

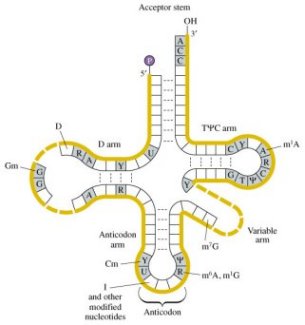
Okay, we have the ribosome at the initiator sequence. What happens next? Well, the initiator sequence (AUG) actually codes for methionine and two different types of tRNA can carry it. One type of tRNA is used in initiation and one type is used in elongation. If you look at the picture above at the top of the picture (labeled acceptor region) the initiator would read ACCA-met-formyl. The anti-codon arm would have 3 G-C pairs in the stem. This is where things really get interesting. Although I mentioned above that the initiator site is AUG, the initiator tRNA can also recognize GUG and UUG. The kicker is that when GUG is used elongation efficiency is reduced by half and when UUG is used elongation is reduced by half again (it would be like if your car ran okay when gas was used as fuel, half as well if beer was used and a quarter as well if water was used - but your car don't work like that does it?). The process is even more variable (that's an important word I will get back to later). I mentioned above that the initiator tRNA has a formyl group on it, but this is actually not necessary, but it does increase the efficiency with which tRNA is bound by a factor (IF-2 for those that know a lot about this) that aids the process. One final wrinkle before I proceed to a description of the process itself. The last two bases in the acceptor stem of the initiator tRNA are unpaired. This prevents the initiator tRNA from being used in elongation. Mutations, however, frequently occur which create a base pair in this position and allow it to function in elongation (in my opinion this undermines everthing Dembski is arguing for and the whole notion of irreducible complexity - but I'm not a biologist so I could be wrong).
Translation


So, shall we build a protein? The top portion of the above picture shows a mRNA strand with an intact ribosome. This is about half a step ahead of where we need to be. Ribosomes are composed of two units - a small and a large which exist in a pool of free ribosomes. Initiation of synthesis is undertaken by the separate subunits. The small subunit binds to the the mRNA and travels to the initiator region. At this point the P-site (see picture above) on the ribosome lies over the initiator region on the mRNA strand forming an initiation complex. The only tRNA that can join the complex, at this point, is the initiator tRNA. The larger subunit joins the complex and makes the A-site available for a tRNA complementary to the second codon on the mRNA. This also is quite variable. Sometimes the process stalls and the the subunits and tRNA are released and the process starts over. Assuming it continues, the tRNA in the P-site transfers it's product to the tRNA in the A-site and the ribosome moves forward one codon. This moves the tRNA in the A-site to the P-site making room for another tRNA in the A-site. The previous occupant of the P-site is released via a third site on the ribosome called the e-site. Here again, there is some variability. Sometimes the wrong tRNA enters the A-site, binds temporarily and is released. Ocassionally, a tRNA enters the A-site with four codons instead of the normal three. Most of the time it is released but occasionally, it's product is incorporated into the protein. At any rate, the ribosome travels down the mRNA strand until it reaches the termination codon, at which point the finished protein is released and the ribosome is released back into the pool. The mRNA usually has more than one ribosome traveling down it and after a while degrades.
The picute below is the real thing. In the bottom portion you can see ribosomes (the round things) traveling down a mRNA strand. Sticking out of the ribosomes are long strings - which are actually proteins in the making ( I don't see any machines, do you?)
Elongation


Summing things up, protein synthesis is a variable process. There are approximately 20,000 ribosomes in cell (and they compose 1/4th of the cell mass). There are approximately 3000 copies of each tRNA as well as 1500 mRNA's in various states of synthesis and decomposition. They all exist in a dynamic equilibrium. To quote Tom Misteli:
"No longer can we think of cellular machines as stable, static, and precisely-assembled complexes, akin to man-made machines."
Instead, researchers found that polymerase subunits came together and formed a complex each time a gene was read, on average every 1.4 seconds. Computer simulations suggest that each formation resulted from random, chaotic interactions between protein subunits that eventually came together in the proper configuration. (emphasis mine) Once a complete polymerase finished reading a gene, the subunits quickly disassembled and scattered throughout the cell. Researchers speculate that the dynamic nature of cellular machines allows components to assemble as needed in response to changing environmental conditions.
All through this post I frequently made comparisons to cars. The point was that designed objects are characterized by their lack of tolerance for variability. I recently put together a book shelf. Problem was, apparently, the manufacturers drill press was out of alignment so the bolt holes didn't match. Which meant I couldn't put the shelf together and so it was completely useless as a book shelf. Cars, motors, planes - anything made by human hands show the same characteristics. Change the operating environment they were designed for (the chronic problems of military hardware in Iraq is a good example) and they rapidly break down. We do not see these qualities in nature though. During protein synthesis any number of things can and do go wrong yet the process continues (it goes without saying that the ability to overcome variability. Phrased another way, biological processes adapt to variability in their environment, designed things don't.
Coming Soon: Part III Semantics and Logic








